- android - 多次调用 OnPrimaryClipChangedListener
- android - 无法更新 RecyclerView 中的 TextView 字段
- android.database.CursorIndexOutOfBoundsException : Index 0 requested, 光标大小为 0
- android - 使用 AppCompat 时,我们是否需要明确指定其 UI 组件(Spinner、EditText)颜色
我需要图像分割方面的帮助。我有脑部肿瘤的 MRI 图像。我需要从 MRI 中移除颅骨(头骨),然后仅分割肿瘤对象。我怎么能在 python 中做到这一点?与图像处理。我试过制作轮廓,但我不知道如何找到并移除最大的轮廓,只得到没有头骨的大脑。非常感谢。
def get_brain(img):
row_size = img.shape[0]
col_size = img.shape[1]
mean = np.mean(img)
std = np.std(img)
img = img - mean
img = img / std
middle = img[int(col_size / 5):int(col_size / 5 * 4), int(row_size / 5):int(row_size / 5 * 4)]
mean = np.mean(middle)
max = np.max(img)
min = np.min(img)
img[img == max] = mean
img[img == min] = mean
kmeans = KMeans(n_clusters=2).fit(np.reshape(middle, [np.prod(middle.shape), 1]))
centers = sorted(kmeans.cluster_centers_.flatten())
threshold = np.mean(centers)
thresh_img = np.where(img < threshold, 1.0, 0.0) # threshold the image
eroded = morphology.erosion(thresh_img, np.ones([3, 3]))
dilation = morphology.dilation(eroded, np.ones([5, 5]))
这些图像与我正在查看的图像相似:
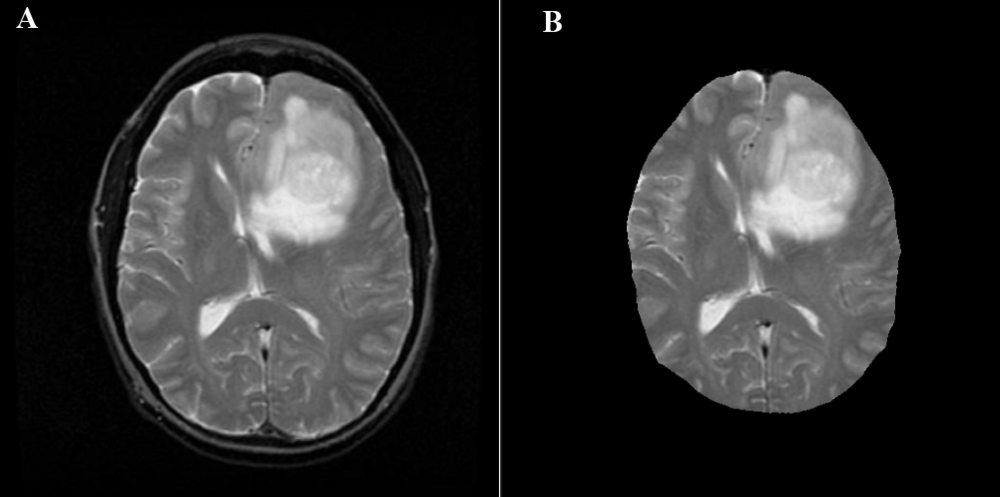
感谢您的回答。
最佳答案
一些初步代码:
%matplotlib inline
import numpy as np
import cv2
from matplotlib import pyplot as plt
from skimage.morphology import extrema
from skimage.morphology import watershed as skwater
def ShowImage(title,img,ctype):
plt.figure(figsize=(10, 10))
if ctype=='bgr':
b,g,r = cv2.split(img) # get b,g,r
rgb_img = cv2.merge([r,g,b]) # switch it to rgb
plt.imshow(rgb_img)
elif ctype=='hsv':
rgb = cv2.cvtColor(img,cv2.COLOR_HSV2RGB)
plt.imshow(rgb)
elif ctype=='gray':
plt.imshow(img,cmap='gray')
elif ctype=='rgb':
plt.imshow(img)
else:
raise Exception("Unknown colour type")
plt.axis('off')
plt.title(title)
plt.show()
作为引用,这是您链接到的大脑+头骨之一:
#Read in image
img = cv2.imread('brain.png')
gray = cv2.cvtColor(img,cv2.COLOR_BGR2GRAY)
ShowImage('Brain with Skull',gray,'gray')
如果图像中的像素可以分为两个不同的强度类别,也就是说,如果它们具有双峰直方图,则Otsu's method可用于将它们阈值化为二进制掩码。让我们检查一下这个假设。
#Make a histogram of the intensities in the grayscale image
plt.hist(gray.ravel(),256)
plt.show()
好的,数据很好地是双峰的。让我们应用阈值,看看我们是怎么做的。
#Threshold the image to binary using Otsu's method
ret, thresh = cv2.threshold(gray,0,255,cv2.THRESH_OTSU)
ShowImage('Applying Otsu',thresh,'gray')
如果我们将蒙版叠加到原始图像上,事情会更容易看
colormask = np.zeros(img.shape, dtype=np.uint8)
colormask[thresh!=0] = np.array((0,0,255))
blended = cv2.addWeighted(img,0.7,colormask,0.1,0)
ShowImage('Blended', blended, 'bgr')
大脑(以红色显示)与面具的重叠非常完美,我们就在这里停下来。为此,让我们提取连接的组件并找到最大的组件,即大脑。
ret, markers = cv2.connectedComponents(thresh)
#Get the area taken by each component. Ignore label 0 since this is the background.
marker_area = [np.sum(markers==m) for m in range(np.max(markers)) if m!=0]
#Get label of largest component by area
largest_component = np.argmax(marker_area)+1 #Add 1 since we dropped zero above
#Get pixels which correspond to the brain
brain_mask = markers==largest_component
brain_out = img.copy()
#In a copy of the original image, clear those pixels that don't correspond to the brain
brain_out[brain_mask==False] = (0,0,0)
ShowImage('Connected Components',brain_out,'rgb')
用你的第二张图片再次运行它会产生一个有很多洞的面具:
我们可以使用 closing transformation 来关闭其中的许多漏洞:
brain_mask = np.uint8(brain_mask)
kernel = np.ones((8,8),np.uint8)
closing = cv2.morphologyEx(brain_mask, cv2.MORPH_CLOSE, kernel)
ShowImage('Closing', closing, 'gray')
我们现在可以提取大脑:
brain_out = img.copy()
#In a copy of the original image, clear those pixels that don't correspond to the brain
brain_out[closing==False] = (0,0,0)
ShowImage('Connected Components',brain_out,'rgb')
如果出于某种原因需要引用它:
Richard Barnes. (2018). Using Otsu's method for skull-brain segmentation (v1.0.1). Zenodo. https://doi.org/10.5281/zenodo.6042312
关于python - MRI(脑肿瘤)图像处理和分割,颅骨去除,我们在Stack Overflow上找到一个类似的问题: https://stackoverflow.com/questions/49834264/
实际上我只需要用JAVA编写一个简单的程序来将MySQL INSERTS行转换为CSV文件(每个mysql表等于一个CSV文件) 在JAVA中使用正则表达式是最好的解决方案吗? 我的主要问题是如何正确
我有一个 txt 文件,其格式为: Key:value Key:value Key:value ... 我想将所有键及其值放入我创建的 hashMap 中。如何让 FileReader(file) 或
已关闭。此问题不符合Stack Overflow guidelines 。目前不接受答案。 要求提供代码的问题必须表现出对所解决问题的最低限度的了解。包括尝试的解决方案、为什么它们不起作用以及预期结果
我每周都会从我的主机下载数据库的备份。它生成一个 .sql 文件,当前大小约为 800mb。此 .sql 文件包含 44 个表。 有什么方法可以通过某些软件将 .sql 文件与所有表分开,以便单独导出
在 iOS 4.0 及更高版本中,有没有一种方法可以在不将整个图像加载到内存的情况下对 CGImage 进行分割?我试图做的是*以编程方式*分割图像,以便在使用大图像的 CATiledLayer 应用
我的 .split() 函数有问题,我有以下字符串: var imageUrl = "Images\Products\randomImage.jpg"; 我想用字符“\”分割,但是,这种情况发生了:
是否可以使用正则表达式将字符串拆分两次?例如,假设我有字符串: example=email@address.com|fname|lname 如何拆分结果为: email@address.com,fna
我正在寻找一种在线程系统(主从)中使用数组的解决方案,它允许我通过用户输入在多个线程上划分矩阵的计算,并将其通过 1 个主线程引导到多个从属线程,这些从属线程计算矩阵的 1 个字段。 我尝试运用我的知
我建立了一个系统来分割包含手写符号的二值图像并对它们进行分类(专门用于音乐)。我知道有商业应用程序可以执行此操作,但这是我尝试将其作为一个项目从头开始。 为了简单起见,假设我的整个图像中有两个元素:
我正在尝试找到一种可接受的复杂性的有效方法 检测图像中的对象,以便将其与周围环境隔离 将该对象分割成它的子部分并标记它们,这样我就可以随意获取它们 我进入图像处理世界已经 3 周了,我已经阅读了很多算
我有一组3D 空间中的点。下图是一个示例: 我想把这些点变成一个面。我只知道点的 X、Y 和 Z 值。例如,查看下图,它显示了从 3D 空间中的点生成的人脸网格。 我在谷歌上搜索了很多,但我找到的是一
我有一个字符串 String placeStr="place1*place2*place3"我想获取包含 place1、place2、place3 的数组,如下所示: String[] places=
我在 Python 中有一个类似于 google.com 的字符串,我想将其分成两部分:google 和 .com。问题是我有一个 URL,例如 subdomain.google.com,我想将其拆分
朋友需要对一个pdf文件进行分割,在网上查了查发现这个pypdf2可以完成这些操作,所以就研究了下这个库,并做一些记录。首先pypdf2是python3版本的,在之前的2版本有一个对应pypdf库。
伙计们,这是一个难以解决的问题,因为它涉及很多硬件细节,所以我想把它放到 EE.SE,但它的主要重点是编程,所以我决定坚持在这里。 我最近怀旧(以及渴望回到 CPU 内在函数),所以我决定自制一个 8
给定 haskell 中的排序列表,我如何获得分段列表,其中连续数字位于同一列表中。例如,如果我有一个排序列表 [1,2,3,4,7,8,10,12,13,15] 结果将是 [[1,2,3 ,4],[
如果我添加三个分割 View ,如下图所示,第三个分割 View (称为 splitView-3)将自动为该分割 View 中的自定义 View 生成约束,例如 customview1 的 Heigh
关闭。此题需要details or clarity 。目前不接受答案。 想要改进这个问题吗?通过 editing this post 添加详细信息并澄清问题. 已关闭 6 年前。 Improve th
如何为馈送给映射器的文件的每一行提供相同文件的拆分? 基本上我想做的是 for each line in file-split { for each line in file{
带有Snappy压缩功能的ORC文件是否可拆分成条形? 据我所知,Snappy Compressed File是不可拆分的。 但我在博客中读到,快速压缩的文件可以在 strip 上拆分。 真的吗? 最

我是一名优秀的程序员,十分优秀!